TLDR:
Boltz-1 marked the beginning of a new era in open-source molecular modeling, introducing an all-atom foundation model capable of predicting the structures of proteins, nucleic acids, and small molecules within a single unified framework widely accessible to everyone. Building on this foundation, Boltz-2 introduced binding affinity prediction, extending beyond structure prediction to capture how tightly molecules recognize and interact with one another. By coupling structure and binding prediction within the same framework, Boltz-2 advanced the understanding of molecular interactions and laid the groundwork for generative design.
Today, we’re excited to announce BoltzGen—an all-atom generative diffusion that can generate new protein binders against any biomolecular target—proteins, nucleic acids, or small molecules alike. Through an extensive experimental campaign conducted with a broad network of collaborators, BoltzGen was tested across an exceptional diversity of targets and use cases. This collective effort pushed the model to its limits and demonstrated unprecedented accuracy, marking a decisive step toward universal binder design.
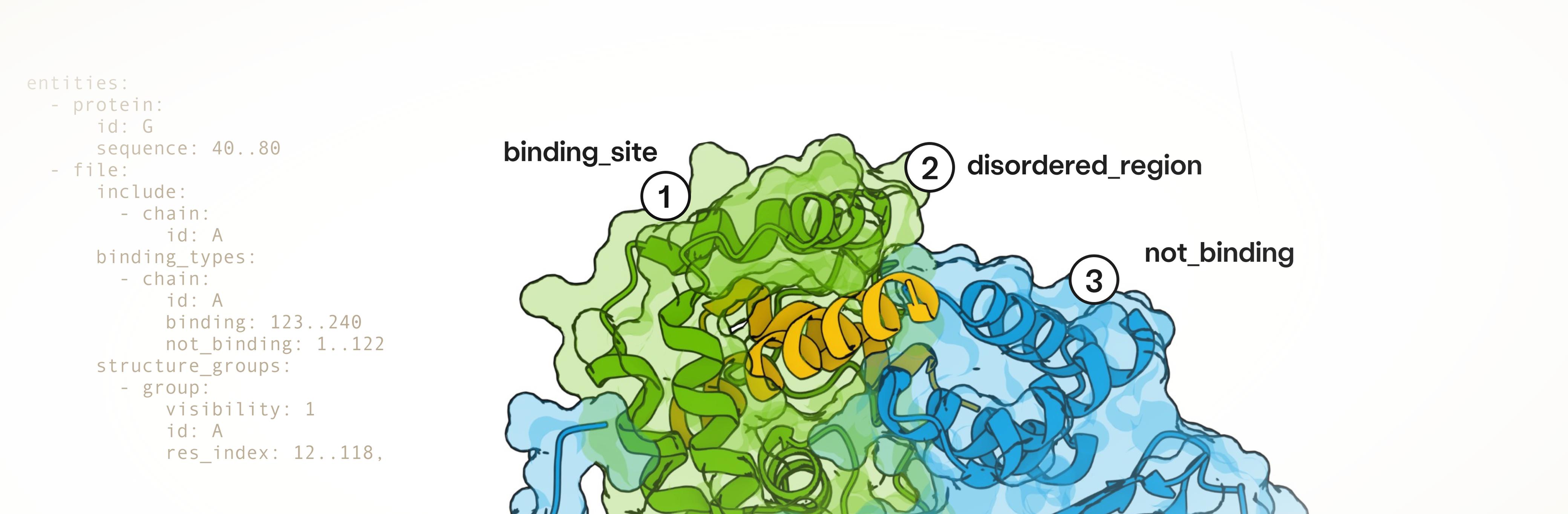
The BoltzGen pipeline uses a single, all-atom generative model that unifies design and structure prediction. A purely geometry-based representation of designed residue types enables scalable training on both tasks simultaneously. As a result, unlike any previous design model, BoltzGen matches the performance of state-of-the-art folding models. BoltzGen's structure-based reasoning about target-binder interactions supports the design of high-affinity binders to novel targets, unrelated to complexes seen during training. This comes with a design specification language that serves as an expressive interface for various constraints – including covalent bonds, structure groups, binding sites, secondary structures and design masks – that steer the diffusion process towards specific design objectives during inference. This language is universal, making it possible to add multiple constraints to a single generation, capturing a wide range of experimental design objectives.
In addition to the core diffusion model, we introduce an integrated pipeline for end-to-end binder design that adds downstream validation, redesign and ranking modules to narrow down a small, diverse set of designs of wet lab-ready designs for experimental screening.
We evaluated BoltzGen on a wide range of difficult benchmarks and real-world binder design challenges in collaboration with leading academic labs and biotechnology companies, validating designs experimentally both for binding and function in living cells. We found strong binders across modalities and biomolecules on targets with high dissimilarity to any bound protein structure. Designs included nanobodies, miniproteins, linear peptides and macrocycle binders against small molecule, peptide, enzyme and protein targets – including experimentally validated binders against intrinsically disordered regions.
BoltzGen was tested on a panel of 9 novel targets with no known binders and less than 30% sequence similarity to any bound molecule or complex in the entire PDB. We chose targets with varied lengths and distinct topologies and folds representing the most challenging and diverse benchmark of any generative protein model to date.

On this challenging benchmark we were able to successfully design nanomolar nanobody binders against 6/9 targets testing 15 designs or fewer per each binder-target pair, demonstrating generalization to unseen regions of interaction space.
To evaluate BoltzGen on real-world biomolecular design problems, we partnered with leading experimental labs and biotech companies to apply the model and pipeline to design and test binders against some of their most challenging targets.

The first set of collaborations yielded experimentally validated binders across a wide range of target types (small molecules, peptides, enzymes, disordered regions) with good affinities and functional readouts including nuclear localization, pathway inhibition and neutralization of microbial activity in live cells.
Like Boltz-1 and Boltz-2, BoltzGen is fully open sourced under an MIT license for unrestricted commercial and academic use so it can be freely used by scientists for real-world problems and applications. BoltzGen is the first of a new generation of all-atom design models and we have only just begun to explore its capabilities. We will be publishing results from our collaborators as they come in and rolling out updates to the model and pipeline as we learn, but we expect the most promising discoveries will come from the community which is why we are committed to making our models accessible to scientists everywhere.
To learn more:
And join us for live presentations, demos, and discussions:
Hannes Stark
On behalf of the BoltzGen team at MIT CSAIL and Jameel Clinic and beyond
